|
Julian McGinnis
I'm a PhD student and researcher in Computer Science at the Munich Centre for Machine Learning (MCML), hosted at the Technical University of Munich (TUM) and TUM Klinikum Rechts der Isar. I work at the intersection of medical imaging, neural fields, and reconstruction algorithms. My research focuses on implicit neural representations with applications to neural compression and super-resolution. I also develop tools for MS lesion segmentation in brain and spinal-cord MRI for Multiple Sclerosis.
|

|
Research (Selected Publications)I'm interested in machine learning, neural compression and signal modeling. Some papers are highlighted. |


|
Optimizing Rank for High-Fidelity Implicit Neural Representations
Julian McGinnis, Florian Hölzl, Suprosanna Shit, Florentin Bieder, Paul Friedrich, Mark Mühlau, Björn Menze, Daniel Rueckert, Benedikt Wiestler Preprint, 2025 project page / arXiv / code / colab We challenge the notion that the low-frequency bias of vanilla MLPs is an architectural limitation, but rather a symptom of stable rank degradation. By using the Muon optimizer to enforce orthogonal updates, we achieve state-of-the-art high-frequency reconstruction with up to 9 dB PSNR improvements. |

|
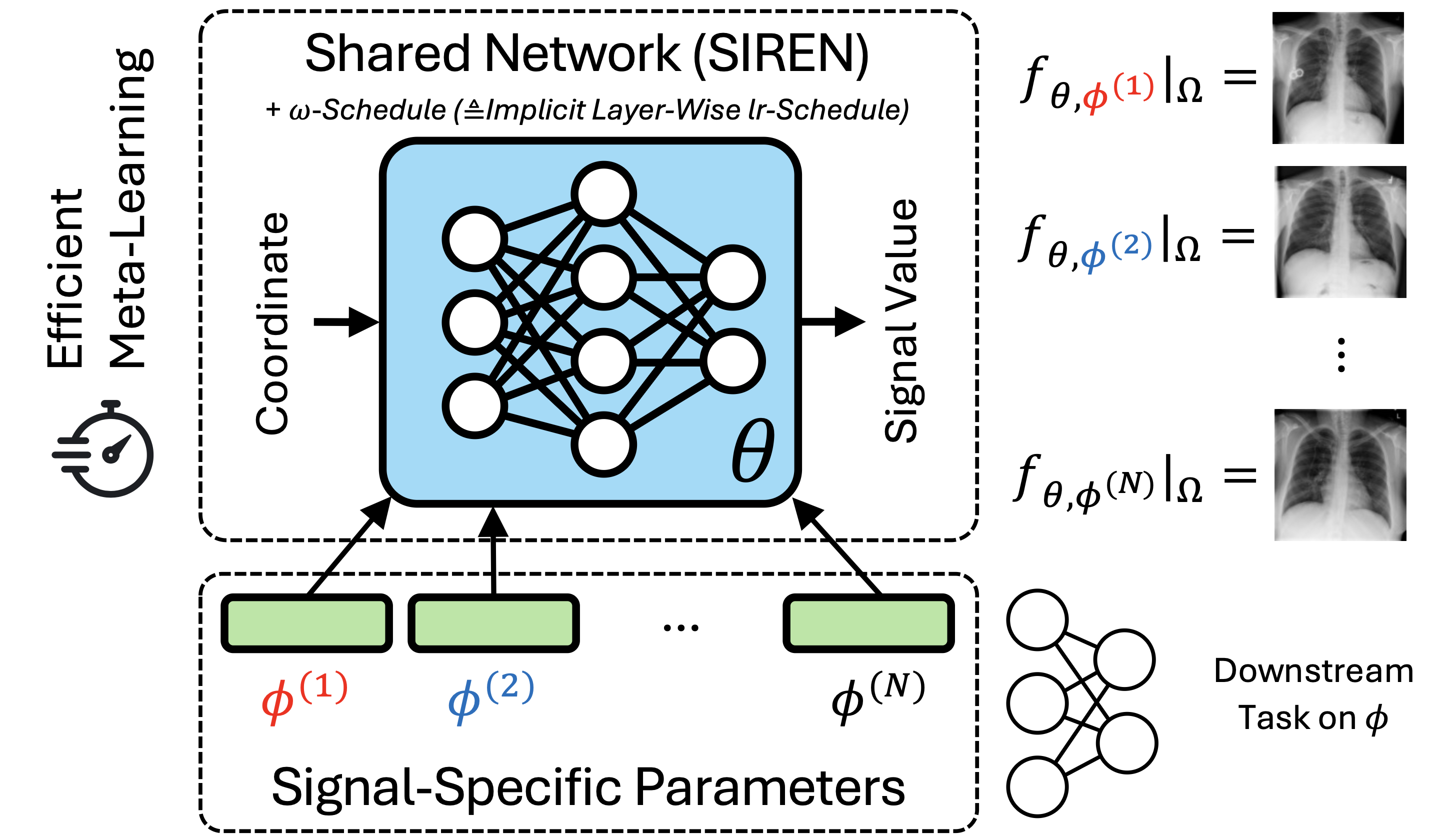
MedFuncta: A Unified Framework for Learning Efficient Medical Neural Fields
Paul Friedrich, Florentin Bieder, Julian McGinnis, Julia Wolleb, Daniel Rueckert, Philippe C. Cattin arXiv, 2025 project page / paper We introduce a scalable meta-learning framework tailored to learning medical neural fields, and provide some theoretical insights showing how SIREN scales learning rates. |

|
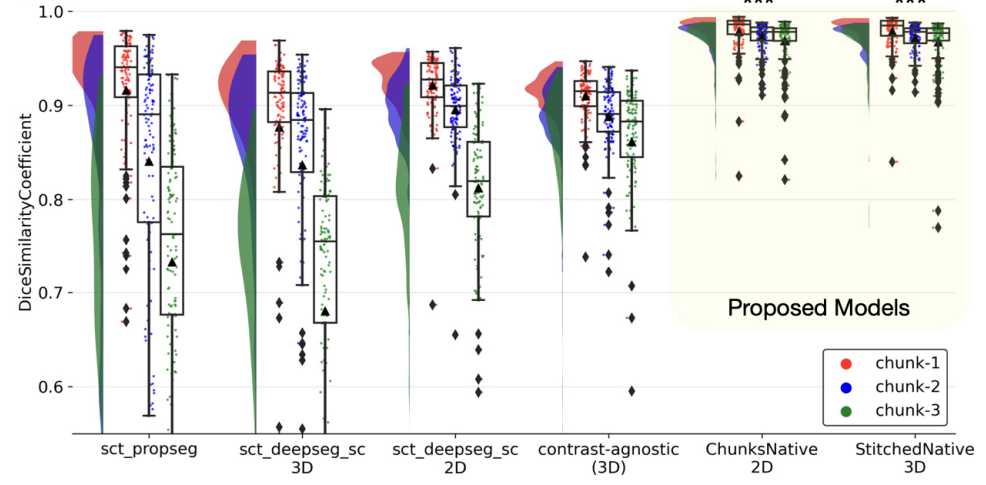
Automatic segmentation of spinal cord lesions in MS: A robust tool for axial T2-weighted MRI scans
Enamundram Naga Karthik*, Julian McGinnis*, Ricarda Wurm, Sebastian Ruehling, Robert Graf, Jan Valosek, Pierre-Louis Benveniste, Markus Lauerer, Jason Talbott, Rohit Bakshi, Shahamat Tauhid, Timothy Shepherd, Achim Berthele, Claus Zimmer, Bernhard Hemmer, Daniel Rueckert, Benedikt Wiestler, Jan S Kirschke, Julien Cohen-Adad, Mark Mühlau Imaging Neuroscience, 2025 code / paper We propose a robust deep learning framework for the automatic segmentation of spinal cord and MS lesions on axial T2-weighted MRI scans. |

|
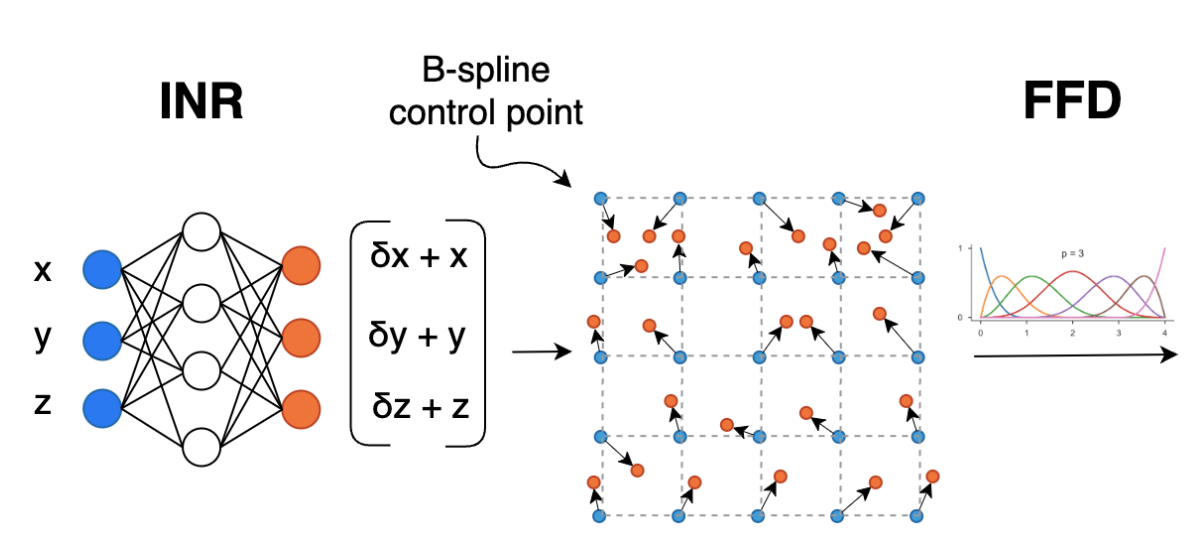
SINR: Spline-enhanced implicit neural representation for multi-modal registration
Vasiliki Sideri-Lampretsa, Julian McGinnis, Huaqi Qiu, Magdalini Paschali, Walter Simson, Daniel Rueckert MIDL, 2024 (Best Paper Award) code / paper We introduce SINR - a method to parameterize the continuous deformable transformation represented by an INR using Free Form Deformations (FFD) enabling robust multi-modal registration while preventing spatial folding. |

|
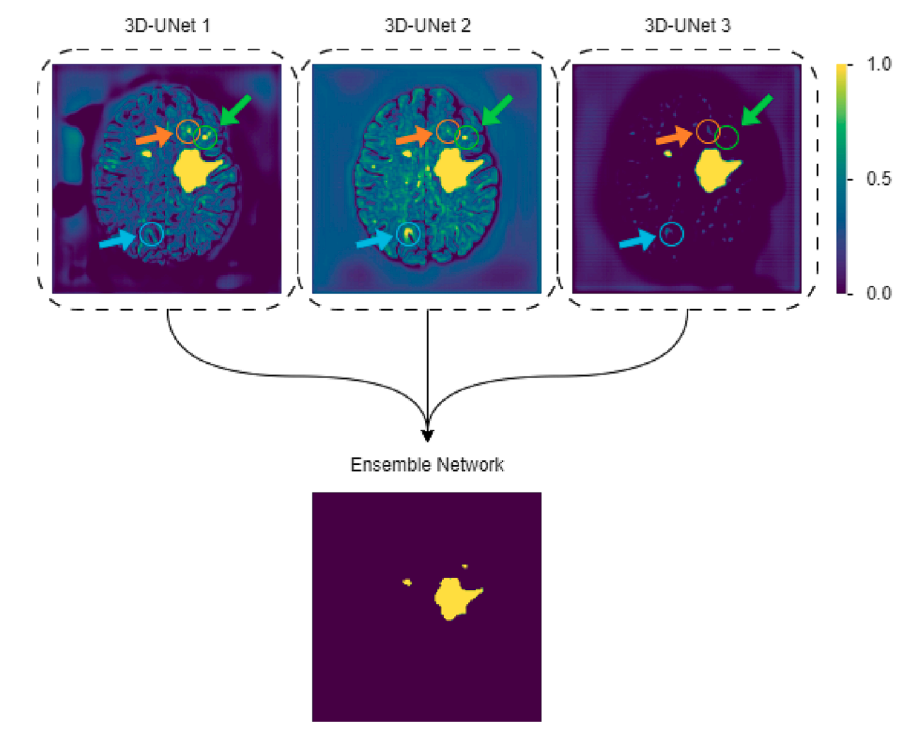
LST-AI: A deep learning ensemble for accurate MS lesion segmentation
Tun Wiltgen, Julian McGinnis, Sarah Schlaeger, Florian Kofler, CuiCi Voon, Achim Berthele, Daria Bischl, Lioba Grundl, Nikolaus Will, Marie Metz, David Schinz, Dominik Sepp, Philipp Prucker, Benita Schmitz-Koep, Claus Zimmer, Bjoern Menze, Daniel Rueckert, Bernhard Hemmer, Jan S. Kirschke, Mark Mühlau, Benedikt Wiestler NeuroImage: Clinical, 2024 (Best Paper Award) code / paper We present LST-AI, an open-source deep learning ensemble for multiple sclerosis lesion segmentation that specifically addresses class imbalance and significantly outperforms traditional tools like LST and standard U-Net architectures. |
|
|
Single-subject Multi-contrast MRI Super-resolution via Implicit Neural Representations
Julian McGinnis* Suprosanna Shit*, Hongwei Bran Li, Vasiliki Sideri-Lampretsa, Robert Graf, Maik Dannecker, Jiazhen Pan, Nil Stolt Ansó, Mark Mühlau, Jan S. Kirschke, Daniel Rueckert, Benedikt Wiestler MICCAI, 2023 code / paper By modeling complementary MRI contrasts in a shared anatomical space with INRs, we are able to super-resolve MRI scans using single subject data alone. |


|
NISF: Neural Implicit Segmentation Functions
Nil Stolt-Ansó, Julian McGinnis, Jiazhen Pan, Kerstin Hammernik, Daniel Rueckert MICCAI, 2023 code / paper We propose a novel family of segmentation models that learn a mapping from a real-valued coordinate space to a shape representation, enabling the segmentation of anatomical shapes in high-dimensional continuous spaces even with sparse or partial data. |

|
Whole Brain Vessel Graphs: A Dataset and Benchmark for Graph Learning and Neuroscience
Johannes C. Paetzold, Julian McGinnis, Suprosanna Shit, Ivan Ezhov, Paul Büschl, Chinmay Prabhakar, Anjany Sekuboyina, Mihail I. Todorov, Georgios Kaissis, Ali Ertürk, Stephan Günnemann, Bjoern H. Menze NeurIPS Datasets and Benchmarks Track, 2021 code / paper We present a biological dataset of whole-brain vessel graphs, now part of Stanford's Open Graph Benchmark Dataset. |
Academic Workshops
 |
|
|
Based on Jon Barron's website. |